Most biomolecules function inside the cell. However, biochemical assays and analytical tools are mainly applied in vitro, in aqueous buffer solutions or crystals. Our research aim is to discover novel biochemical function by studying biomolecular reactions directly in cells under health and disease conditions. To analyze biomolecular reactions in vivo with high spatio-temporal resolution, we developed novel in-cell spectroscopic and microscopic techniques that range from temperature-induced relaxation to single molecule spectroscopy. We apply the methods in environments of increasing complexity ranging from cytomimetic media and bacteria to (neuronal) cells and multicellular organisms. Our current research interest lies in the field of neurodegenerative diseases with a focus on the following research topics.
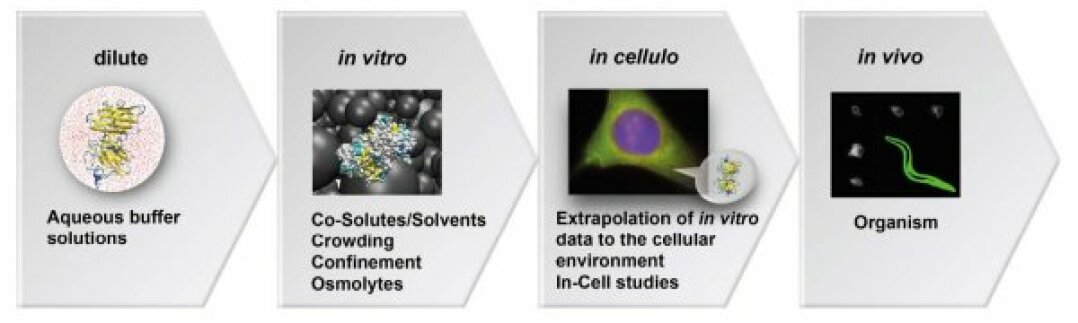
Most methods to measure the rates of protein folding and aggregation of disease-related proteins rely on in vitro methods such as ThT binging assays. Using experiments in cytomimetic media, our lab elaborated a novel thermodynamic model to understand how different compounds like ions[1], metabolies[2,3], crowders[4], drugs[5] or dehydration[6] could modify the underlying equilibria. Using a biosensor framework, we could interpret in-cell folding experiments under physiological conditions[7-10], cell stress[6,9,11,12] or differentiation[12]. These insights led to a novel classification of disease-related mutants[2,3,13,14]. We further implemented a novel assay to measure protein aggregation kinetics with high spatio-temporal resolution directly in cells[5,13,15] that can be used to screen novel classes of aggregation inhibitors as potential drugs[5,16]. In a DFG project with Prof. K. Huber (Paderborn), we now decipher the different driving forces that govern in-cell aggregation kinetics.
Molecular chaperones constitute a cellular mechanism to prevent aggregation and to retain the proteome folded in space and time. In a HFSP project we collaborated to establish a novel method to measure proteostasis capacity under various cellular conditions[9,10]. This work led us to investigate the different mechanisms of chaperones to modify the folding, misfolding and aggregation rates of different protein substrates.
It is now clear that biomolecular condensates play a profound role in folding, aggregation and amyloid-associated pathologies. Our current work will advance the understanding of functional and dysfunctional condensates by revealing how stress granules, in conjunction with other factors like molecular chaperones, could reshape folding and aggregation pathways to assist protein homeostasis, or conversely, how the integrity of SGs could be endangered by sequestration of different amyloidogenic species. In the first funding period of DFG-SPP2191, we could show that early folding events play a role in the sequestration of proteins[14,17] such as pathogenic mutants of the enzyme SOD1[14]. In the second funding period, we will extend our studies to analyze the folding and aggregation pathways in the stress granules in direct comparison to the cytoplasm which will be possible by confocal Fast Relaxation Imaging a method recently devolved in our lab.
Metabolic enzymes can be spatially and temporally organized in cells forming regulatory multi-enzyme complexes called metabolons. Little is known about such complexes since they cannot be studied by disruptive protocols in vitro. Based on our previous work in which we learned how enzymatic stability and catalysis changes in cytomimetic and cytoplasmic environments compared to dilute solution[3,18-20], we investigate the stability and metabolic activity of enzymes in metabolons (GRK 2223: Protein Complex Assembly).
Compared to proteins, little is known about RNA structure and folding in cells. Our lab was first to measure the stability of well-folded RNA hairpins inside cells[8]. In our current research, we focus on disease-relevant RNA sequences like the CAG triplet-repeat RNA hairpin involved in Huntington’s disease[21].
[1] Senske, M. et al. The temperature dependence of the Hofmeister series: thermodynamic fingerprints of cosolute–protein interactions. Phys. Chem. Chem. Phys. 18, 29698–29708 (2016).
[2] Gnutt, D. et al. Stability Effect of Quinary Interactions Reversed by Single Point Mutations. J. Am. Chem. Soc. 141, 4660–4669 (2019).
[3] Brylski, O. et al. Cellular ATP Levels Determine the Stability of a Nucleotide Kinase. Front. Mol. Biosci. 8, 1259 (2021).
[4] Gao, M. et al. Modulation of human IAPP fibrillation: cosolutes, crowders and chaperones. Phys. Chem. Chem. Phys. 17, 8338–8348 (2015).
[5] Vöpel, T. et al. Inhibition of Huntingtin Exon-1 Aggregation by the Molecular Tweezer CLR01. J. Am. Chem. Soc. 139, 5640–5643 (2017).
[6] Gnutt, D. et al. Imperfect crowding adaptation of mammalian cells towards osmotic stress and its modulation by osmolytes. Mol. BioSyst. 13, 2218–2221 (2017).
[7] Gnutt, D. et al. Excluded-Volume Effects in Living Cells. Angew. Chem. Int. Ed. 54, 2548–2551 (2015).
[8] Gao, M. et al. RNA Hairpin Folding in the Crowded Cell. Angew. Chem. Int. Ed. 55, 3224–3228 (2016).
[9] Wood, R. J. et al. A biosensor-based framework to measure latent proteostasis capacity. Nat Commun 9, 287 (2018).
[10] Raeburn, C. B. et al. A biosensor of protein foldedness identifies increased “holdase” activity of chaperones in the nucleus following increased cytosolic protein aggregation. J. Biol. Chem. 298, 102158 (2022).
[11] Hämisch, B. et al. Self‐Assembly of Pseudo‐Isocyanine Chloride as a Sensor for Macromolecular Crowding In Vitro and In Vivo. Chem. Eur. J. 26, 7041–7050 (2020).
[12] Gnutt, D. et al. Protein Folding Modulation in Cells Subject to Differentiation and Stress. Front. Mol. Biosci. 6, 38 (2019).
[13] Brylski, O. et al. Disease-Related Protein Variants of the Highly Conserved Enzyme PAPSS2 Show Marginal Stability and Aggregation in Cells. Front. Mol. Biosci. 9, (2022).
[14] Samanta, N. et al. Sequestration of Proteins in Stress Granules Relies on the In-Cell but Not the In Vitro Folding Stability. J. Am. Chem. Soc. 143, 19909–19918 (2021).
[15] Büning, S. et al. Conformational dynamics and self-association of intrinsically disordered Huntingtin exon 1 in cells. Phys. Chem. Chem. Phys. 19, 10738–10747 (2017).
[16] Azoulay-Ginsburg, S. et al. Chemical chaperones targeted to the endoplasmic reticulum (ER) and lysosome prevented neurodegeneration in a C9orf72 repeat expansion drosophila amyotrophic lateral sclerosis (ALS) model. Pharmacol. Rep (2021).
[17] Ribeiro, S. et al. Protein folding and quinary interactions: creating cellular organisation through functional disorder. FEBS Letters 592, 3040–3053 (2018).
[18] Dirkmann, M. et al. A Multiperspective Approach to Solvent Regulation of Enzymatic Activity: HMG-CoA Reductase. ChemBioChem 19, 153–158 (2018).
[19] Dhar, A. et al. Structure, function, and folding of phosphoglycerate kinase are strongly perturbed by macromolecular crowding. Proc. Natl. Acad. Sci. 107, 17586–17591 (2010).
[20] Ganguly, A. et al. Elucidation of the Catalytic Mechanism of a Miniature Zinc Finger Hydrolase. J. Phys. Chem. B 121, 6390–6398 (2017).
[21] Hautke, A. C. et al. S. Folding Stability and Self-Association of a Triplet-Repeat (CAG)20 RNA Hairpin in Cytomimetic Media. ChemSystemsChem 3, e2000052 (2021).
You are leaving our website now. Please note that this link opens an external website. This may be subject to less strict privacy policies and may collect personal information. Further information on the protection of your data can be found in our privacy policy